PNAS
3D printing of microscopic bacterial communities
J. L. Connell, E. T. Ritschdorff, M. Whiteley, and J. B. Shear
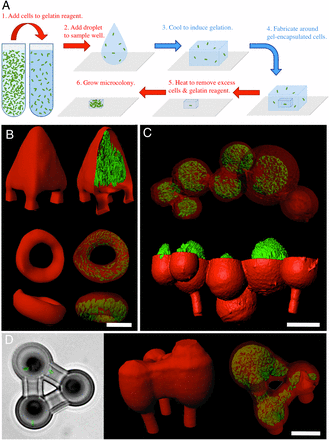
Bacteria communicate via short-range physical and chemical signals, interactions known to mediate quorum sensing, sporulation, and other adaptive phenotypes. Although most in vitro studies examine bacterial properties averaged over large populations, the levels of key molecular determinants of bacterial fitness and pathogenicity (e.g., oxygen, quorum-sensing signals) may vary over micrometer scales within small, dense cellular aggregates believed to play key roles in disease transmission. A detailed understanding of how cell–cell interactions contribute to pathogenicity in natural, complex environments will require a new level of control in constructing more relevant cellular models for assessing bacterial phenotypes. Here, we describe a microscopic three-dimensional (3D) printing strategy that enables multiple populations of bacteria to be organized within essentially any 3D geometry, including adjacent, nested, and free-floating colonies. In this laser-based lithographic technique, microscopic containers are formed around selected bacteria suspended in gelatin via focal cross-linking of polypeptide molecules. After excess reagent is removed, trapped bacteria are localized within sealed cavities formed by the cross-linked gelatin, a highly porous material that supports rapid growth of fully enclosed cellular populations and readily transmits numerous biologically active species, including polypeptides, antibiotics, and quorum-sensing signals. Using this approach, we show that a picoliter-volume aggregate of Staphylococcus aureus can display substantial resistance to β-lactam antibiotics by enclosure within a shell composed of Pseudomonas aeruginosa.